łŪļĻ•–°ľ§Ōarrows§ÚĽ»Õ—§∑§∆…Ń≤Ť§∑§Ņ°£őőįŤ§Ōrect§«…Ń≤Ť§∑°Ędensity§«…Ń≤Ť•Ņ•§•◊§ÚĽōńͧ«§≠§Ž°£
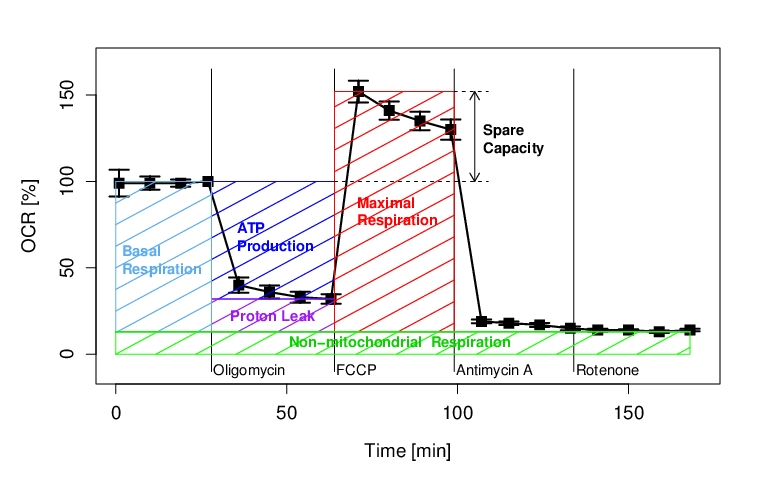
Ōņ łļÓņģĽĢ§őR§«§ő•į•ť•’…Ń≤Ťő„§Ú§§§Į§ń§ęĶů§≤§Ž°£•ř•¶•Ļ§«ī √Ī§ňĹ٧Ī§ §§§ő§¨∑ÁŇņ§«§Ę§Ž°£
łŪļĻ•–°ľ§Ōarrows§ÚĽ»Õ—§∑§∆…Ń≤Ť§∑§Ņ°£őőįŤ§Ōrect§«…Ń≤Ť§∑°Ędensity§«…Ń≤Ť•Ņ•§•◊§ÚĽōńͧ«§≠§Ž°£

cls <- function(){ while(dev.cur() > 1) {dev.off()} }
cls()
tp <- read.table("r2-20150611.csv", sep=",", header=T)
par(pin=c(7, 4))
par(mai=c(1,1,0.5,0.5)) $ BLUR
par(ps=14)
plot(0,0,type="n",xlim=range(1:170),ylim=range(-10:170),xlab="Time [min]",ylab="OCR [%]")
lines(tp[,"TIME"], tp[,"Control"], lwd=2, lty=1)
points(tp[,"TIME"], tp[,"Control"], lwd=1, pch=15, cex=1.5)
arrows(tp[,"TIME"], tp[,"Control"] + tp[,"Cdev"], tp[,"TIME"], tp[,"Control"] - tp[,"Cdev"], angle = 90, length = 0.1, lwd=2)
arrows(tp[,"TIME"], tp[,"Control"] - tp[,"Cdev"], tp[,"TIME"], tp[,"Control"] + tp[,"Cdev"], angle = 90, length = 0.1, lwd=2)
lines(c(28,28), c(-10,165), lwd=1, lty=1, )
lines(c(64,64), c(-10,165), lwd=1, lty=1, )
lines(c(99,99), c(-10,165), lwd=1, lty=1, )
lines(c(134,134), c(-10,165), lwd=1, lty=1, )
rect(28, 32, 64,100, col="blue", density=4)
text(35, 65, "ATP\nProduction", pos=4, cex=0.8, offset=0.1, col="blue", fg="white", font=2)
rect(28, 13, 64, 32, col="purple", density=4)
text(33, 21, "Proton Leak", pos=4, cex=0.8, offset=0.1, col="purple", font=2)
rect(0, 13, 28, 100, col="steelblue2", density=4)
text(0, 52, " Basal\n Respiration", pos=4, cex=0.8, offset=0.1, col="steelblue2", font=2)
rect(64, 13, 99, 152, col="red", density=4)
text(70, 80, "Maximal\nRespiration", pos=4, cex=0.8, offset=0.1, col="red", font=2)
lines(c(0,168), c(13,13), lwd=1.5, lty=1, col="green3")
rect(0,0, 168,13, col="green", density=4)
text(50, 6, "Non-mitochondrial Respiration", pos=4, cex=0.8, offset=0.1, col="green3", font=2)
lines(c(99,109), c(152,152), lty=2)
lines(c(64,109), c(100,100), lty=2)
arrows(105, 100, 105, 152, code=3, length=0.1)
text(107, 122, "Spare\nCapacity", pos=4, cex=0.8, offset=0.1, col="black", font=2)
text(28, -10, "Oligomycin", pos=4, cex=0.8, offset=0.1)
text(64, -10, "FCCP", pos=4, cex=0.8, offset=0.1)
text(99, -10, "Antimycin A", pos=4, cex=0.8, offset=0.1)
text(134,-10, "Rotenone", pos=4, cex=0.8, offset=0.1)
dev.copy2eps(device=postscript, file="r2-XF24_SF1b.eps", width=8, height=5, family="Helvetica")
cls()
cls <- function(){ while(dev.cur() > 1) {dev.off()} }
cls()
tp <- read.table("r2-XF24_1.csv", sep=",", header=T)
par(pin=c(7, 4))
par(mai=c(1,1,0.5,0.5)) $ BLUR
par(ps=16)
plot(0,0,type="n",xlim=range(1:170),ylim=range(0:200),xlab="Time [min]",ylab="OCR [%]")
lines(tp[,"TIME"], tp[,"Control"], lwd=2, lty=1, col="lightskyblue")
lines(tp[,"TIME"], tp[,"H0.1mM"], lwd=2, lty=2, col="royalblue1")
lines(tp[,"TIME"], tp[,"H0.3mM"], lwd=2, lty=3, col="slateblue3")
lines(tp[,"TIME"], tp[,"H1mM"], lwd=2, lty=4, col="orchid")
points(tp[,"TIME"], tp[,"Control"], lwd=1, pch=15, cex=1.5, col="lightskyblue")
points(tp[,"TIME"], tp[,"H0.1mM"], lwd=1, pch=16, cex=1.5, col="royalblue1")
points(tp[,"TIME"], tp[,"H0.3mM"], lwd=1, pch=17, cex=1.5, col="slateblue3")
points(tp[,"TIME"], tp[,"H1mM"], lwd=1, pch=18, cex=1.5, col="orchid")
arrows(tp[,"TIME"], tp[,"Control"] + tp[,"Cdev"], tp[,"TIME"], tp[,"Control"] - tp[,"Cdev"], angle = 90, length = 0.1, lwd=2, col="lightskyblue")
arrows(tp[,"TIME"], tp[,"Control"] - tp[,"Cdev"], tp[,"TIME"], tp[,"Control"] + tp[,"Cdev"], angle = 90, length = 0.1, lwd=2, col="lightskyblue")
arrows(tp[,"TIME"], tp[,"H0.1mM"] + tp[,"H0.1mMdev"], tp[,"TIME"], tp[,"H0.1mM"] - tp[,"H0.1mMdev"], angle = 90, length = 0.1, lwd=2, col="royalblue1")
arrows(tp[,"TIME"], tp[,"H0.1mM"] - tp[,"H0.1mMdev"], tp[,"TIME"], tp[,"H0.1mM"] + tp[,"H0.1mMdev"], angle = 90, length = 0.1, lwd=2, col="royalblue1")
arrows(tp[,"TIME"], tp[,"H0.3mM"] + tp[,"H0.3mMdev"], tp[,"TIME"], tp[,"H0.3mM"] - tp[,"H0.3mMdev"], angle = 90, length = 0.1, lwd=2, col="slateblue3")
arrows(tp[,"TIME"], tp[,"H0.3mM"] - tp[,"H0.3mMdev"], tp[,"TIME"], tp[,"H0.3mM"] + tp[,"H0.3mMdev"], angle = 90, length = 0.1, lwd=2, col="slateblue3")
arrows(tp[,"TIME"], tp[,"H1mM"] + tp[,"H1mMdev"], tp[,"TIME"], tp[,"H1mM"] - tp[,"H1mMdev"], angle = 90, length = 0.1, lwd=2, col="orchid")
arrows(tp[,"TIME"], tp[,"H1mM"] - tp[,"H1mMdev"], tp[,"TIME"], tp[,"H1mM"] + tp[,"H1mMdev"], angle = 90, length = 0.1, lwd=2, col="orchid")
lines(c(28,28), c(0,200), lwd=1, lty=1, )
lines(c(64,64), c(0,200), lwd=1, lty=1, )
lines(c(99,99), c(0,200), lwd=1, lty=1, )
lines(c(134,134), c(0,200), lwd=1, lty=1, )
text(28,0, "Oligomycin", pos=4, cex=0.8, offset=0.1)
text(64,0, "FCCP", pos=4, cex=0.8, offset=0.1)
text(99,0, "Antimycin", pos=4, cex=0.8, offset=0.1)
text(134,0, "Rotenone", pos=4, cex=0.8, offset=0.1)
leg <- c(" Control",
expression(paste(" ", paste(paste(H[2],O[2])," 0.1 mM 1hr"))),
expression(paste(" ", paste(paste(H[2],O[2])," 0.3 mM 1hr"))),
expression(paste(" ", paste(paste(H[2],O[2])," 1.0 mM 1hr"))))
legend("topright", legend = leg, pch = c(15,16,17,18), lty = c(1,2,3,4), bg="white", y.intersp=2, xjust=1, col=c("lightskyblue","royalblue1","slateblue3","orchid"))
dev.copy2eps(device=postscript, file="r2-XF24_F5a_col.eps", width=8, height=5, family="Helvetica")
cls()
cls <- function(){ while(dev.cur() > 1) {dev.off()} }
cls()
par(pin=c(4, 4))
par(mai=c(1,1,0.5,0.5)) $ BLUR
par(ps=15)
leg2 <- c(" Control", "0.1 mM", "0.3 mM", "1.0 mM")
jc1 <- c(1.0079774, 0.957234643, 0.931015671, 0.886195603)
jc1_dev <- c(0.021088766, 0.01139586, 0.014554344, 0.024889494)
C <- barplot(jc1, ylim=c(0.8,1.099), ylab="RFU(590nm)/RFU(535nm)", space=1, col=c("lightskyblue","royalblue1","slateblue3","orchid"),names.arg=leg2, xpd=F)
arrows(C, jc1 - jc1_dev, C, jc1 + jc1_dev, angle = 90, length = 0.1)
arrows(C, jc1 + jc1_dev, C, jc1 - jc1_dev, angle = 90, length = 0.1)
box("plot",lty=1)
drawbraces <- function(x1,x2,y) {
segments(x1,y, x2,y)
segments(x1,y, x1,y-0.005)
segments(x2,y, x2,y-0.005)
}
drawbraces(1.5,3.5,1.05)
drawbraces(1.5,5.5,1.065)
drawbraces(1.5,7.5,1.08)
text(2.5,1.051, "*", pos=4, cex=0.9, offset=0)
text(3.5,1.066, "*", pos=4, cex=0.9, offset=0)
text(4.5,1.081, "*", pos=4, cex=0.9, offset=0)
dev.copy2eps(device=postscript, file="r2-XF24_F4_color.eps", width=5, height=5, family="Helvetica")
cls()
…Ń≤Ť żłĢ§Ú§“§√§Į§Í ÷§Ļ§ő§Ōylim§«•—•ť•Š°ľ•Ņ§ÚĶ’§ň§Ļ§ž§–ő…§§°£par(new=T)§«ĹŇ§Õ…Ń§≠§Ļ§ž§–őĺ żłĢ§ő•į•ť•’§¨§«§≠§Ž°£ ľī§ő…Ń≤Ťľę¬ő§Ōaxis§«§Ę§Ž§¨°Ę•—•ť•Š°ľ•Ņ§Úpar()§ę§ťľŤ∆ņ§∑§Ņ§Í§Ļ§Ž§ő§Ō√ő§ť§ §§§»§š§šŐŐŇ›§«§Ę§Ž°£
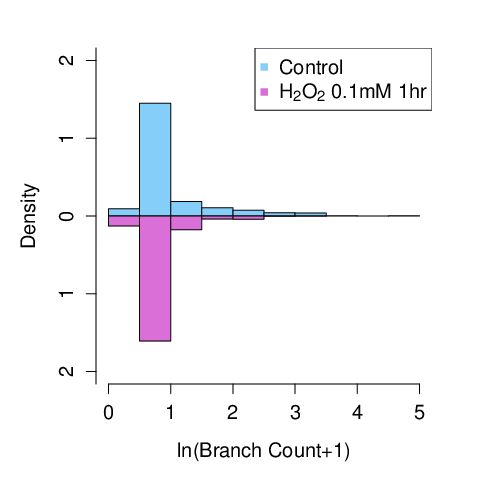
cls <- function(){ while(dev.cur() > 1) {dev.off()} }
cls()
tp <- read.table("r2-20160406_H1_MT_combined.csv", sep=",", header=T)
str(tp)
par(pin=c(4, 4))
par(mai=c(1,1,0.5,0.5)) $ BLUR
par(ps=15)
c1 <- subset(tp,TYPE=="CONT")[["Branches"]]
c2 <- subset(tp,TYPE=="HYD" )[["Branches"]]
plot.new()
plot.window(ylim = c(-2, 2), xlim = c(0, 5))
par(new = TRUE)
hist(log(c1+1),probability=TRUE,xlim=c(0,5),ylim=c(-2,2),col="lightskyblue",axes = FALSE, xlab = "", ylab = "", main = "",breaks=8)
par(new = TRUE)
hist(log(c2+1),probability=TRUE,xlim=c(0,5),ylim=c(2,-2),col="orchid",axes = FALSE, xlab = "ln(Branch Count+1)", ylab = "Density", main = "",breaks=5)
axis(side = 2, at = pretty(par()$usr[3:4]), labels = abs(pretty(par()$usr[3:4])))
axis(side = 1)
leg <- c(" Control",
expression(paste(" ", paste(paste(H[2],O[2])," 0.1mM 1hr "))))
legend("topright", legend = leg, pch = c(15,15), bg="white", y.intersp=2, xjust=1, col=c("lightskyblue","orchid"))
dev.copy2eps(device=postscript, file="r2-XF24_F3_branch.eps", width=5, height=5, family="Helvetica")
cls()
<ubajas> Technically, my first language was Turbo Pascal, but I
started over with Perl 10 years later (not having
programmed in the meantime). I'm obviously damaged goods.
<iank> ubajas: heh, I read that as "I started with (perl 10)
(years later)" instead of "I started with perl (10 years
later)" :)
<rindolf> Perl 10!
<rindolf> Perl for the Fourth Millenium.
<jagerman> I thought Perl 6 was supposed to be timeless
<ubajas> iank: Maybe I should have added a comma. :-]
<jagerman> Perl ‚ąě
<iank> perl6 has existed since the beginning of time, or at least
it will have existed since then once $Larry finds a time
machine.
<simcop2387> iank: i'm sorry but larry is the prophet i am the
messanger! i will be the one to take it back!
<iank> WHAT.
<simcop2387> iank: its MY TIME MACHINE!
* iank smacks simcop2387 around
<jagerman> iank: So it'll be like that Star Trek episode, where they
say that the development of computers are caused by time
travel from the future?
<jagerman> Except that they were too stupid (like most Voyager
writers) to get their facts right, and thought computers
started in the 70s
-- The Messiah of Perl
-- #perl, Freenode
What happened to Christopher Michael Pilato?
Is he gone?
Is he gone for good?
Is he gone for better?
Is he gone for best?
Is he gone forever?
Will he return?
Who is Christopher Michael Pilato, anyway?
Shlomi Fish
-- Shlomi Fish
-- Adapted from an IRC Monologue